6 The pipeline
- RStudio Environment. Let’s rearrange the panel layout:
Tools>Global Options…
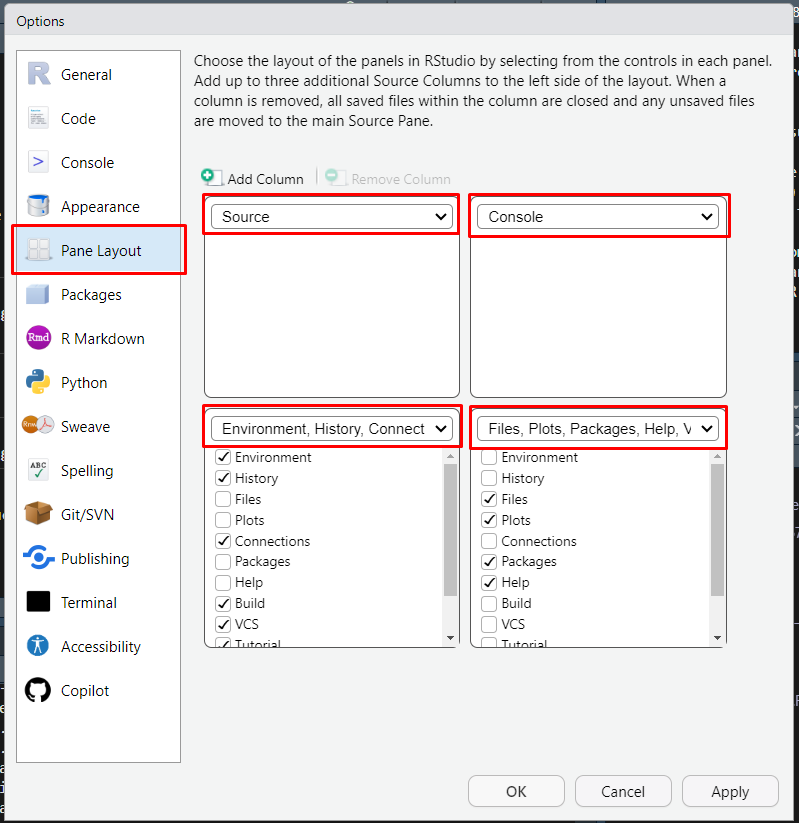
Pane Layout
6.1 Essential data management and folder structure
├── config.R
├── data_derived
│ ├── Australia_SDG_14.csv
│ ├── sdg_14.csv
│ ├── sdg_14_unclos_map.csv
│ └── sdg_3_1_2.csv
├── data_provided
│ ├── country-to-region-mapping.csv
│ ├── Ocean Accounts Diagnostic Tool_formatted.pdf
│ ├── SDG-DSD-Guidelines.pdf
│ ├── SDG.xlsx
│ └── SDG_Updateinfo.xlsx
├── DatSciTrain_SDGs_API_R.Rproj
├── figures_and_tables
│ ├── fig2.png
│ └── sdg14_Australia.docx
├── LICENSE
├── R
│ ├── do_clean.R
│ ├── do_get_sdg_api.R
│ ├── do_map.R
│ ├── do_plot.R
│ └── do_tab_Australia.R
├── README.md
└── run.R6.2 config.R
# packages
if (!require(data.table)) {
install.packages("data.table")
library(data.table)
}
if (!require(ggplot2)) {
install.packages("ggplot2")
library(ggplot2)
}
if (!require(sf)) {
install.packages("sf")
library(sf)
}
if (!require(RColorBrewer)) {
install.packages("RColorBrewer")
library(RColorBrewer)
}
if (!require(rnaturalearth)) {
install.packages("rnaturalearth")
library(rnaturalearth)
}
if (!require(rnaturalearthdata)) {
install.packages("rnaturalearthdata")
library(rnaturalearthdata)
}
## set folder names
folder_names <- c("data_derived", "data_provided", "figures_and_tables")
for (folder_name in folder_names) {
if (!dir.exists(folder_name)) {
dir.create(folder_name)
cat("Folder", folder_name, "created.\n")
} else {
cat("Folder", folder_name, "already exists.\n")
}
}
## source functions
file_list <- list.files(path = "R", pattern = "\\.R$", full.names = TRUE)
# Source each .R file
for (file in file_list) {
source(file)
}6.3 run.R
source("config.R")
### 1. Download ####
# Use the function to download SDGs data
do_get_sdg_api()
### 2. Data cleaning ####
# Function to clean the data downloaded
indat <- do_clean()
### 3. Tabulating ####
tab <- do_tab_country(indat, country = "Indonesia")
### 4. Visualise ####
# Generate and interactive plot with the data cleaned
do_plot()
### 5. Map ####
do_map()6.4 do_get_sdg_api
do_get_sdg_api <- function(
output = "data_derived/sdg_14.csv"
){
# (Client URL) command line tool that enables data exchange between a device
# and a server through a terminal
curl <- paste0(
'curl -X POST --header "Content-Type: application/x-www-form-urlencoded" ',
'--header "Accept: application/octet-stream" ',
'-d "goal=14" ',
'"https://unstats.un.org/sdgapi/v1/sdg/Goal/DataCSV" -o',
output)
# Execute cURL
system(curl)
}6.5 do_clean
do_clean <- function() {
# options(scipen = 1000)
# Load data
indat <- fread(file.path("data_derived", "sdg_14.csv"))
# mapping <- fread(file.path("data_provided", "country-to-region-mapping.csv"))
# Keep only the values that are either blank or 'A' under 'Observation Status', drop the rest
indat <- indat[`[Observation Status]` == "" | `[Observation Status]` == "A"]
# Replace '-' with '_' across all disaggregation values
cols_to_replace <- grep("\\[.*\\]",
names(indat),
value = TRUE)
indat[, (cols_to_replace) := lapply(.SD, function(x) gsub("-", "_", x)),
.SDcols = cols_to_replace]
return(indat)
}6.6 do_tab_country
do_tab_country <- function(
indat,
country
){
# Filter the input data for the specified country
foo <- indat[GeoAreaName == country]
# Select specific columns from the filtered data
foo14 <- foo[, .(Indicator,
SeriesDescription,
TimePeriod,
Source)]
# Convert TimePeriod column to numeric for easier calculations
foo14[, NumericTimePeriod := as.numeric(TimePeriod)]
# Calculate min and max year for each SeriesDescription using TimePeriod
time_ranges <- foo14[, .(
StartYear = min(NumericTimePeriod, na.rm = TRUE),
EndYear = max(NumericTimePeriod, na.rm = TRUE)
), by = SeriesDescription]
# Create a time range string (e.g., "2000-2020" or "2000" if start and end year are the same)
time_ranges[, TimeRange := ifelse(StartYear == EndYear, as.character(StartYear), paste(StartYear, EndYear, sep = "-"))]
# Merge the new time range back to the main data.table
foo14 <- merge(foo14, time_ranges, by = "SeriesDescription", all.x = TRUE)
# Drop temporary columns that are no longer needed
foo14[, NumericTimePeriod := NULL]
foo14[, StartYear := NULL]
foo14[, EndYear := NULL]
foo14[, TimePeriod := NULL]
# Keep unique rows based on SeriesDescription
unq <- unique(foo14, by = "SeriesDescription")
# Select the final columns to include in the output
unq <- unq[, .(Indicator, SeriesDescription, TimeRange, Source)]
# Define the output file name based on the country
out_name <- paste0("data_derived/", country, "_SDG_14.csv")
# Write the data to a CSV file
fwrite(unq, out_name)
return(unq)
}6.7 do_plot
do_plot <- function(){
# Subset the data to only include rows where the Indicator is "14.7.1"
sdg1471 <- indat[Indicator=="14.7.1"]
# Order the subsetted data by GeoAreaName in ascending order
sdg1471 <- sdg1471[order(sdg1471$GeoAreaName, decreasing = FALSE)]
# Display the unique GeoAreaNames in the subsetted and ordered data
unique(sdg1471$GeoAreaName)
# Further subset the data to only include rows where the GeoAreaName is "Indonesia"
sdg1471_ind <- sdg1471[GeoAreaName=="Indonesia"]
# Let's make a simple plot using base R
plot(
sdg1471_ind$TimePeriod,
sdg1471_ind$Value
)
# Some improvements:
# type = "l"
# col = "blue"
# lwd = 2
# main = "Sustainable Fisheries as a proportion of GDP in Indonesia"
# xlab = "Year"
# ylab = "(%)"
# Comparing Indonesia with other countries, subsetting first
sdg1471_comp <- sdg1471[GeoAreaName %in% c("Indonesia", "Malaysia", "Cook Islands")]
# Create the plot using ggplot2
ggplot(sdg1471_comp,
aes(x = TimePeriod, y = Value, color = GeoAreaName, group = GeoAreaName)) +
geom_line(size = 1.2) +
labs(title = "Sustainable Fisheries as a proportion of GDP",
x = "Year",
y = "(%)",
color = "Country") +
theme_minimal()
# Include world averages for comparison
sdg1471_comp_two <- sdg1471[GeoAreaName %in% c("Indonesia", "Malaysia", "Cook Islands", "World")]
# Create the plot using ggplot2
p <- ggplot(sdg1471_comp_two,
aes(x = TimePeriod, y = Value, color = GeoAreaName, group = GeoAreaName)) +
geom_line(size = 1.2) +
labs(title = "Sustainable Fisheries as a proportion of GDP (including World average)",
x = "Year",
y = "(%)",
color = "Country") +
theme_minimal()
return(p)
}6.8 do_map
do_map <- function()
{
# United Nations Convention on the Law of the Sea (UNCLOS)
# Indicator 14.c.1: Number of countries making progress in ratifying,
# accepting and implementing through legal, policy and institutional frameworks,
# ocean-related instruments that implement international law, as reflected in
# the United Nations Convention on the Law of the Sea, for the conservation and
# sustainable use of the oceans and their resources
foo <- indat[SeriesCode == "ER_UNCLOS_RATACC" | SeriesCode == "ER_UNCLOS_IMPLE"]
foo1 <- foo[SeriesCode == "ER_UNCLOS_RATACC", .SD[which.max(as.numeric(TimePeriod))], by = GeoAreaName]
foo2<- foo[SeriesCode == "ER_UNCLOS_IMPLE", .SD[which.max(as.numeric(TimePeriod))], by = GeoAreaName]
# Get country polygons
world <- rnaturalearth::ne_countries(scale = "medium", returnclass = "sf")
setnames(foo1, "GeoAreaName", "name")
setnames(foo2, "GeoAreaName", "name")
foo2_names <- unique(foo2$name)
foo1_names <- unique(foo1$name)
world_names <- unique(world$name)
names_diff_foo2_world <- setdiff(foo2_names, world_names)
names_diff_foo1_world <- setdiff(foo1_names, world_names)
print("Names in foo2 not in world:")
print(names_diff_foo2_world)
print(names_diff_foo1_world)
foo2[name == "Republic of Korea", name := "South Korea"]
foo2[name == "United Kingdom of Great Britain and Northern Ireland", name := "United Kingdom"]
foo2[name == "Russian Federation", name := "Russia"]
foo2[name == "Venezuela (Bolivarian Republic of)", name := "Venezuela"]
foo1[name == "Republic of Korea", name := "South Korea"]
foo1[name == "United Kingdom of Great Britain and Northern Ireland", name := "United Kingdom"]
foo1[name == "Russian Federation", name := "Russia"]
foo1[name == "Venezuela (Bolivarian Republic of)", name := "Venezuela"]
foo2_imple <- foo2[SeriesCode == "ER_UNCLOS_IMPLE"]
foo1_rat <- foo1[SeriesCode == "ER_UNCLOS_RATACC"]
foo3 <- rbind(foo2_imple, foo1_rat)
foo3_map <- merge(world, foo3, by = "name", all.x = TRUE, fill = TRUE)
setDT(foo3_map)
# Replace NaN and NA in 'Value' with NA for uniform handling
foo3_map[, Value := fifelse(is.nan(Value) | is.na(Value), as.numeric(NA), Value)]
foo3_map[, ValueFactor := cut(Value, breaks = c(0, 50, 69, 79, 89, 100),
include.lowest = TRUE, right = TRUE,
labels = c("0-50%", "51-69%", "70-79%", "80-89%", "90-100%"))]
foo4 <- foo3_map[!is.na(foo3_map$Value)]
foo5 <- foo4[, c("name", "Value", "Indicator", "TimePeriod", "SeriesDescription"), drop = FALSE]
write.csv(foo5, "data_derived/sdg_14_unclos_map.csv", row.names = FALSE)
foo3_map <- st_as_sf(foo3_map)
equal_earth_projection <- st_crs("+proj=eqearth +datum=WGS84")
p <- ggplot(data = foo3_map) +
geom_sf(aes(geometry = geometry, fill = ValueFactor), color = "white", size = 0.2) +
scale_fill_brewer(palette = "YlGnBu", name = "", na.value = "grey") +
coord_sf(crs = equal_earth_projection, datum = NA) + # Apply Equal Earth projection
theme(
panel.background = element_rect(fill = "white"),
legend.position = "top"
)
ggsave("figures_and_tables/fig_map.png", plot = p, width = 10, height = 6, dpi = 300, units = "in")
}